
Research Director: Shigeyuki Yokoyama
(Professor, Graduate School of Science, The University of Tokyo / Chief Scientist, RIKEN)
Research Term: 1996-2001
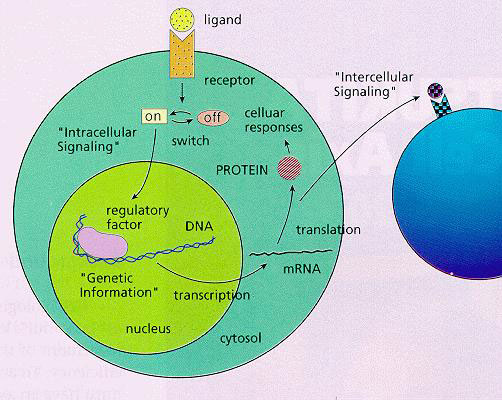
The Yokoyama CytoLogic project investigated the cell’s information-processing system to learn more about how it works, how it can be modified and how its features can be used in artificial information processing systems.
Research Results
DNA and RNA: Many unnatural base pairs were designed to have exclusive pairing specificities, minimally interacting with four natural bases. They were chemically synthesized and their specificities (in the replication of DNA and transcription of DNA to RNA) were analysed through in vitro analysis.
Among them, an unnatural base pair of 2-amino-6-(N,N- dimethylamino)purine (x) and pyridin-2-one (y) was found to undergo specific transcription. The ribonucleoside triphosphate of y is selectively incorporated into RNA opposite to x in the template by T7 RNA polymerase. The results enable RNA biosynthesis for the specific incorporation of unnatural nucleotides to the desired positions. Further, another base pair, 2-amino-6-(2-thieny1)purine (s) and y, was obtained.
Modified tRNA having both an artificial anticodon and an unnatural aminoacyl moiety was prepared. The artificial base was introduced to the DNA of a cell signaling protein. Transcription of this DNA followed by translation with the modified tRNA gave an artificial protein that had the unnatural amino acid site specifically.
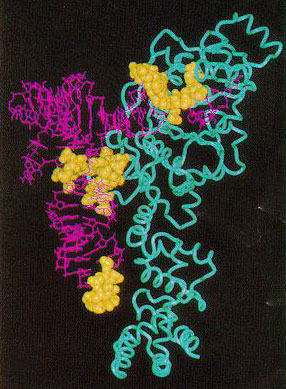
tRNA: With the intention of obtaining variant tRNAs which are useful for the translation of an artificial codon and the incorporation of the corresponding unnatural amino acid, a method to make variant tRNAs which have both an artificial anticodon and new identity determinants was established.
Aminoacyl-tRNA synthetase: An in vivo selection method of aminoacyl-tRNA synthetase variants was developed, and E. coli Arg-tRNA synthetase variants that aminoacylate an amber suppressor tRNA with anticodon CUA were isolated from random mutant pools. The recognition mechanism was clarified.
Based on the 3-D structure of the complex of Tyr-tRNA synthetase and tyrosine, variants of Tyr-tRNA synthetase, which can incorporate an unnatural tyrosine analog (3-iodotyrosine) more than the natural amino acid tyrosine, were prepared.
EGF receptor signaling: The unnatural tyrosine analog (3-iodotyrosine) was site-specifically incorporated, in response to the UAG or s-containing codon, into proteins in vitro and / or in vivo (eukaryotic cells). This finding is useful for the formation of an artificial signaling network. Actually, the adaptor Grb2 SH2 domain, which binds to the phophotyrosine site in the EGF receptor, was engineered to bind preferentially to the phospho-3- iodotyrosine site.
RNA aptamers: RNA aptamers that specifically bind to the Ras-binding domain of Raf-1 were isolated from a random-sequence RNA pool. The anti-Raf-1 aptamers may be useful for clarifying the complicated signaling network and the formation of an artificial network downstream of tyrosine kinases such as the EGF receptor.
RNA aptamers that bind to and inhibit a ribosome-inactivating protein, pepocin, were isolated from a random-sequence RNA pool. This enabled making mutants of E. coli which can survive after the incorporation of an unnatural base.
tRNA and aminoacyl-tRNA synthetase
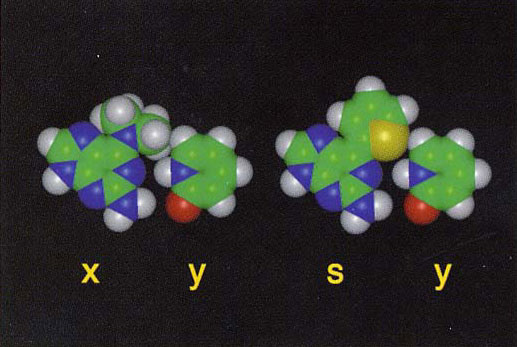
Unnatural x-y and s-y base pairs
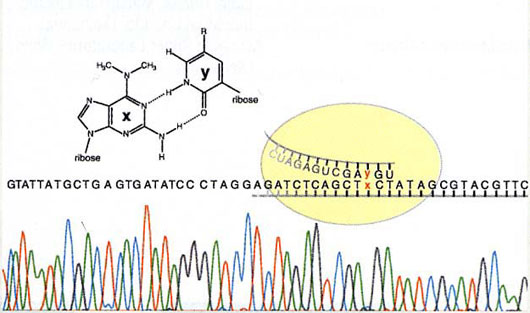
The x-y base pair for specific transcription and DNA sequencing