Research Director: Keiichi Namba
(Professor, Graduate School of Frontier Biosciences, Osaka University)
Research Term: 1997-2002
Namba Protonic NanoMachine Project studied the structure and function of the bacterial flagellum as a huge molecular system based on the organization and movement of the individual atoms that build it.
Research Results
Crystallization of flagellar proteins: To determine the molecular structures of flagellin to identify the mechanism of the mechanical switch of sub-Å precision and of many other flagellar proteins to understand how they work, they were cloned, overproduced, purified and crystallized. For fiber-forming proteins, it was first necessary to produce fragments that do not polymerize by removing the terminal regions that stabilize the fiber structure.
X-ray crystallography: Atomic models of fragments of flagellin, hook protein, and HAP3 were built at around 2 Å resolution from diffraction data obtained by using brilliant X-rays from SPring-8 beam-lines. Crystals of other flagellar proteins yielded high-resolution diffraction data as well. The a-axis molecular array in the flagellin crystal was identified to be the flagellar protofilament with the shorter repeat distance of its two distinct conformations, whose repeat distance is 51.9 Å and 52.7 Å. Docking of the molecular array into a density map of the flagellar filament obtained by electron cryomicroscopy also proved it.
Mechanical switch: A computer simulation of the protofilament extension was carried out to find the flagellin structure responsible for the high-precision mechanical switch. This work greatly stimulated the research of the mechanical properties of proteins.
Electron cryomicroscopy and image analysis: Electron cryomicroscopy and image analysis were used to analyze the flagellar filament or the cap-filament complex. Thousands of images were collected and carefully processed, one by one, and either helical image reconstruction or single-particle image analysis was used to reconstruct the three-dimensional images at high-resolution.
Flagellar cap as assembly nanomachine: Flagellin subunits are arranged in a helical manner to form the filament with approximately 5.5-fold symmetry, but the cap bound at its distal end is a HAP2 pentamer with 5-fold symmetry, having a pentagonal table top with five leg domains. The molecular structure revealed a rotary mechanism of the cap to promote self-assembly of flagellin to the filament-end, which is achieved by its binding over the symmetry-mismatch.
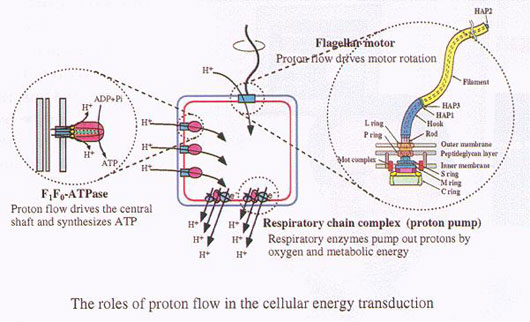
Structure analysis of motor: Structural studies of the motor protein complexes were also conducted by electron cryomicroscopy and X-ray crystallography to understand how the motor produces the torque by proton flow even at an energy level comparable to the thermal noise, and why the bushing works well without lubrication. Motor proteins having membrane-spanning structures were overproduced and new purification methods were developed.
Flagellar motor rotation: To understand the mechanism of the highly efficient proton motor, the rotation speed and its fluctuation or stepping motion were examined by nanophotometry. A new optical system incorporating a specially designed high-sensitivity quadrant photo-sensor was developed for measuring single flagellar motor rotation at high temporal and spatial resolution. Data indicate that the motor rotates with unexpectedly large, rapid fluctuations and even stops occasionally for several milliseconds.