Progress Report
Understanding and Control of Virus-Human Interaction Networks[3] Development of analysis frameworks for imaging and math analysis to comprehend Virus-Human Interaction Networks.
Progress until FY2024
1. Outline of the project
To prevent severe disease progression following viral infections, it is essential to understand host response networks during the pre-disease phase and enable ultra-early prediction and intervention. This project has established a range of in vitro and in vivo infection models—including for SARS-CoV-2, influenza, RSV, arboviruses, hemorrhagic fever viruses, and chronic hepatitis viruses—and visualized and quantified host immune responses and disease-related mechanisms using omics and advanced imaging technologies.
In FY2024, we integrated the resulting high-dimensional datasets with mathematical and computational approaches to build a unified framework for classifying diverse disease trajectories. By strengthening our collaboration with clinical data sources, we are also accelerating the translation of findings from animal models into clinical contexts. Ultimately, the project aims to clarify how infection outcomes diverge—between mild and severe cases—and to generate insights that contribute to future infectious disease preparedness and precision medicine.
2. Outcome so far
The imaging group has developed technologies for visualizing host responses to viral infection at single-cell resolution in organoids, tissues, and whole organisms. Using light-sheet and super-resolution microscopy, they enabled dynamic 3D imaging of viral spread and immune activity. Spatial transcriptomic methods, such as multiplex RNA-FISH and MERSCOPE, revealed inflammation-related molecular patterns, while electron microscopy visualized viral particles and neural markers in astrocytes.
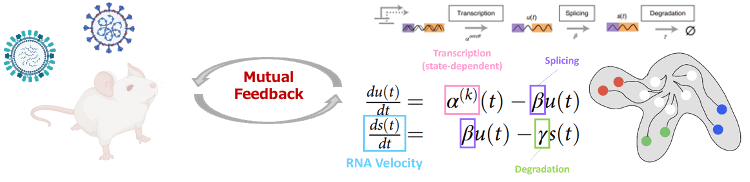
The mathematical modeling group analyzed high-dimensional data from multiple viral infection models to identify shared therapeutic targets and build models for disease stratification. Integration with clinical data improved prediction of early biomarkers and understanding of pre-disease states. For mpox, they developed a PCR-based simulator that estimates transmission risk, allowing safe shortening of isolation periods by up to one week and supporting the development of flexible, evidence-based guidelines.
3. Future plans
We will integrate high-resolution imaging, omics, and AI-based modeling to stratify disease states and improve early prediction. Using organoids and animal models, we aim to capture immune dynamics and disease progression in real time, advancing personalized medicine and enabling cross-viral insights with strong translational potential.