Progress Report
Understanding and Control of Virus-Human Interaction Networks[2] Analysis of Host Response Network
Progress until FY202
1. Outline of the project
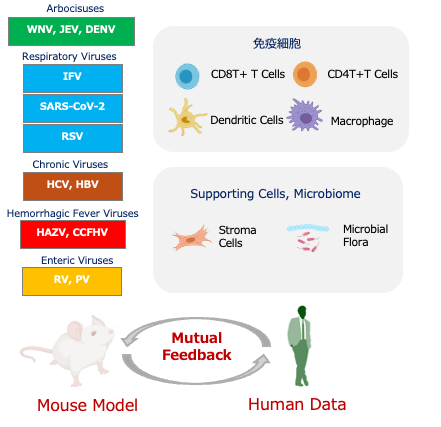
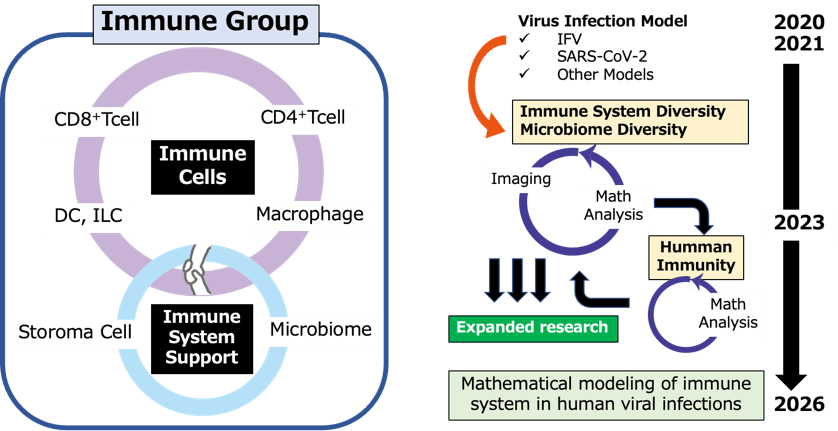
Using various viral infection models, we conducted single-cell analyses of immune cells and, with mathematical and imaging researchers, visualized response patterns and developed models. In FY2024, we identified key molecules linked to age- and disease-specific immune responses, including those in macrophages and neutrophils, and showed that chemokine regulation and gut microbiota shifts relate to disease severity. Human sample analysis revealed age-related immune changes and post-vaccination differences, helping bridge findings with animal models. These efforts support personalized prevention and new treatment strategies.
2. Outcome so far
In FY2024, studies using mouse and human samples clarified how immune cells and gut microbiota respond to viral infections and influence disease severity. Age, host constitution, and pre-infection cellular or microbiota states were found to significantly impact infection dynamics and outcomes. These insights support early risk prediction and the development of personalized, pre-symptomatic interventions.
Researchers further identified key roles for group 2 innate lymphoid cells (ILC2s) in antiviral defense. ILC2s were shown to be activated at infection sites and to regulate immune responses via interactions with epithelial and stromal cells.
These findings provide a basis for exploring how human conditions such as allergy, aging, and obesity modulate responses to viral infection.
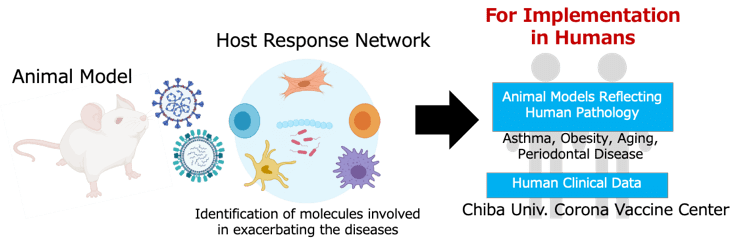
3. Future plans
Going forward, we will further integrate data from animal models and human samples to uncover the molecular mechanisms underlying individual differences in response to viral infections. By visualizing and modeling immune, microbiota, and support cell networks over time, we aim to enhance the accuracy of early risk prediction. These efforts will ultimately support the development of personalized preventive strategies and innovative therapeutic approaches that allow for intervention before symptoms appear.