Progress Report
Understanding and Control of Virus-Human Interaction Networks[1] Analysis of virus infection networks
Progress until FY2024
1. Outline of the project
We have developed diverse in vitro and in vivo models for viral infections and analyzed host response networks using omics approaches. Collaborating with experts in immunology, mathematical modeling, and imaging, we are identifying early biomarkers and therapeutic targets for severe disease.
Using models for viruses such as SARS-CoV-2, RSV, influenza, and others, we aim to uncover shared host responses and establish a new disease classification framework. Ongoing integration with clinical data supports the translational application of these findings.
Objectives
- To establish infection models that reflect human disease and evaluate them through collaboration with Themes 2 and 3.
- To clarify mechanisms underlying the progression to severe disease.
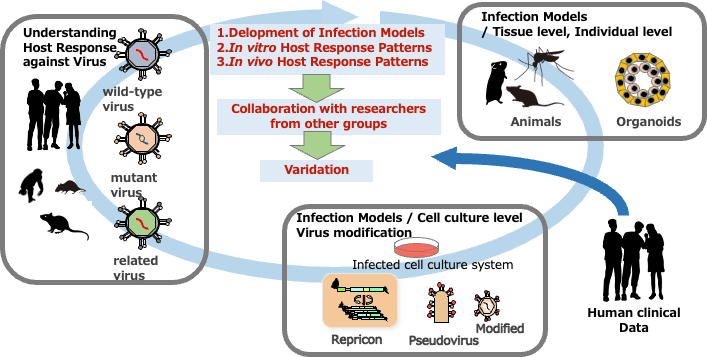
2. Outcome so far
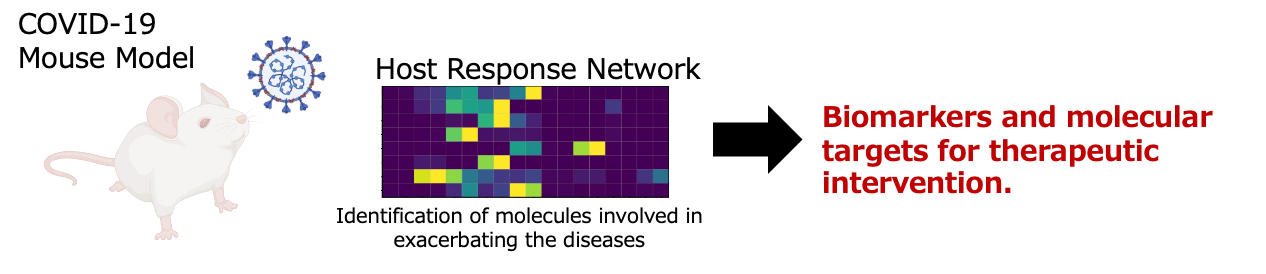
In FY2024, we refined virus infection models for severe respiratory diseases and advanced the extraction of host response networks using comprehensive omics analyses. In a SARS-CoV-2 animal model, we identified host response pathways and predictive biomarkers for severe disease. Comparative analysis with human clinical data revealed candidate molecules potentially involved in pathogenesis. We also expanded infection models for other viruses, including influenza and RSV, to explore shared mechanisms among respiratory viruses.
By FY2024, infection models for enteric, arthropod-borne, hemorrhagic, and persistent viruses were completed. We are now collaborating with R&D teams 2 and 3 to extract host response networks. Focusing on PBMCs, which are available across all models, we have begun cross-model sample collection for comparative analysis.
3. Future plans
Using the various virus infection models established by FY2024, we will advance comparative analyses of host response networks derived from omics data. By identifying common biomarkers and predictive indicators, we aim to enable ultra-early prediction of disease severity and the discovery of therapeutic targets. Integration with human clinical data will further accelerate the clinical application of our findings.