Progress Report
Challenge toward the Control of Intractable Cancer through Understanding of Molecular, Cellular, and Interorgan Networks[3] Technology development for the creation of innovative diagnostic and treatment concepts based on understanding of the onset process of cancer
Progress until FY2024
1. Outline of the project
In this item, cell biology will explore the role of candidates of early diagnosis markers and therapeutic targets revealed from the multi-hierarchical data of intractable cancer patients. We are developing the experimental system and technology necessary to evaluate its validity.
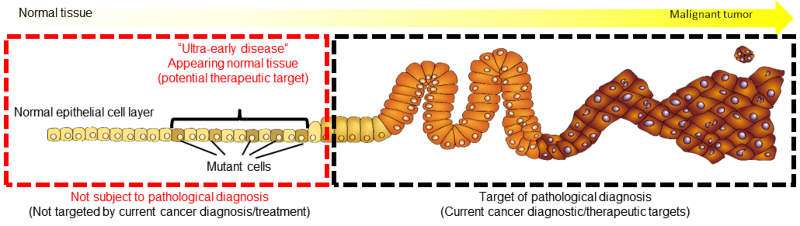
Based on mouse models, normal tissues have multiple mechanisms to eliminate abnormal cells caused by various cellular stresses (cell senescence, bacterial infection, genetic mutation, etc.) to prevent the occurrence of cancer. In addition, changes in metabolism, stem cells, cell adhesion and morphology are central characteristics of cancer cells. It is necessary to know when these cancer development process occur.
In addition to model mice and cancer cell lines, we use clinical specimens containing early-stage lesions, and patient organoids as experimental systems. We are working to understand the process and create new diagnostic and treatment concepts.
2. Outcome so far
Network analysis focusing on cell competition and cell senescence
We identified a molecule whose expression increases in the pancreas of a cell competition model mouse. This molecule was expressed in premalignant lesions (ADM) of pancreatic carcinoma in model mice and human clinical samples.
A genetic screen in Drosophila identified a group of genes involved in the elimination of cancer mutant or normal cells.
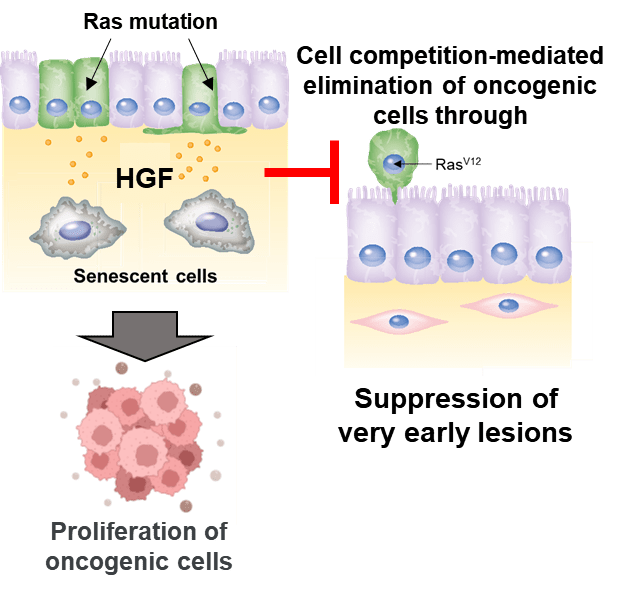
Mutant cells were found to be eliminated from normal cell populations through cell competition by a “kick-me-out” signal. We also revealed that mitochondrial fatty acid oxidation initiated by nuclear DNA damage leads to cellular senescence.

We found that elimination of cancer mutant cells by cell competition is suppressed by senescent cells and senolytic drugs are effective in eliminating cancer mutant cells. We developed a fluorescent probe that targets DPP4 to visualize senescent cells.

Network analysis focusing on stem cells, cell polarity, and epithelial-to-mesenchymal transition
Through experiments and mathematical models, we found that glycolysis is asymmetrically distributed via aPKC during asymmetric division of cancer stem cell, resulting in cancer heterogeneity.

Using a mouse model with organoids, we found that an mTOR inhibitor is promising treatment for refractory ovarian cancer.

We found that Wnt5b-Ror1 signaling promotes the proliferation of pancreatic cancer, and that pancreatic cancer patients with high Ror1 expression have a poor prognosis.

Network analysis focusing on intestinal flora, cancer immunity, and metabolism
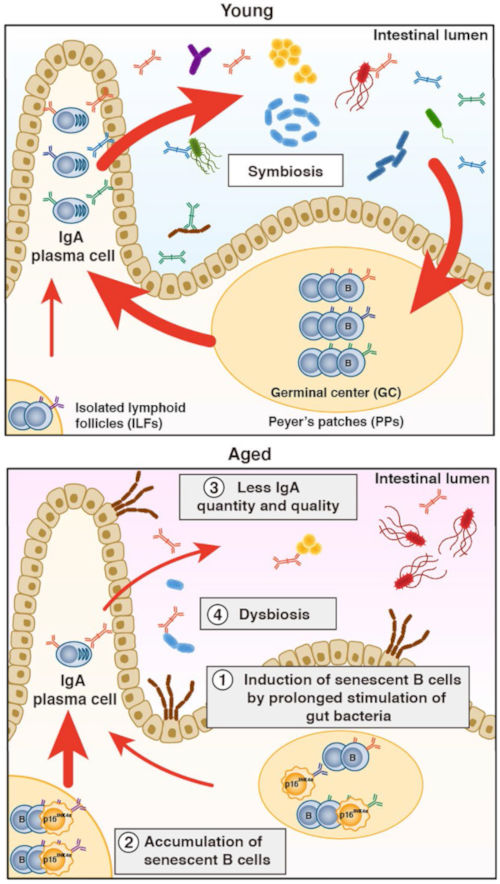
Using a model mouse in which cancer development increases due to the accumulation of senescent cells, we identified intestinal bacteria that promote the accumulation of senescent cells and the development of cancer. Furthermore, we discovered that age-related aging of B cells causes disturbances in the intestinal flora.
We analyzed stresses in the tumor microenvironment that induce the expression of cancer-specific antigens through RNA metabolism and developed simultaneous measurement system for newly synthesized RNA and translation.

We have established a system to detect metabolic changes of amino acids and keto acids in vivo in real time and identified the metabolic pathway of branched-chain amino acids in cancer cells. We found that an inhibitor against chromatin-related factors is effective for leukemia.
3. Future plans
For early diagnostic markers, we will verify the molecules whose expression is increased in pancreatic precancerous lesions and will advance research to understand the precancerous states and the onset of cancer using organoids.
We will also address issues such as cell competition, the relationship between cellular senescence and the intestinal microbiota, metabolic shifts in cancer, changes in stem cell characteristics and in cell polarity.