Research Results
With a bucket of water
Developing an innovative survey method to identify fish species FY2017

- Michio Kondoh (Professor, Ryukoku University)
- CREST
- Establishment of core technology for the preservation and regeneration of marine biodiversity and ecosystems "Application of environmental DNA for qualitative monitoring of fish community and ecosystem assessment" Research Director (2013-2018)
Identifying species of fish from DNA released by the fish in seawater
It is important to reveal biological diversity for conserving regional biota and managing biological resources. Waters around Japan have the highest level of fish diversity, and deepening the understanding of fish fauna is indispensable for their conservation and management. However, conventional methods for fish biota surveys that utilize visual observation and fishing gears require assistance from many experts with advanced knowledge, in addition to a large amount of labor and long-term research.
As a complementary method to solve these problems, "environmental DNA metabarcoding" has come under the spotlight. This method makes it possible to identify species of fish present at a specific area through extracting and analyzing DNA collected from elements including excrement and mucus in seawater. In "Strategic Basic Research Program" (CREST), JST , research and development on advanced measurement technologies for understanding marine biodiversity and ecosystems and on models contributing to future prediction are being conducted. In this research program, a research team including Masaki Miya (Head of Department of Ecology and Environmental Sciences, Natural History Museum and Institute, Chiba), Toshifumi Minamoto (Specially-appointed Professor, Kobe University) and Reiji Masuda (Associate Professor, Kyoto University) led by Professor Micho Kondoh is working on the development of a new monitoring method to analyze fish-derived DNA contained in seawater and to promptly and quantitatively identify the species composition, biomass and genetic characteristics of fish.
Entering a big data age with new technology possessing higher detectability
In recent years, it was revealed that DNA released with their body surface mucus and excrement outside the body of living organisms is floating in seawater. This is called "environmental DNA." Currently, DNA can be easily read out like "bar codes on goods," and the species of fish can be detected from the read out information (DNA base sequence).
Metabarcoding, in which water in the surveyed water area is collected and then environmental DNA in the water is examined to detect fish species living in that area, has greatly reduced the labor and time involved in fish biota surveys. At the same time, this method has made it possible to conduct surveys on fish pieces at multiple points in a short period of time, which was impossible with the conventional survey methods. Currently, research using this innovative field survey method is conducted in the sea and rivers all over the world. However, due to the low detectability of metabarcoding, survey research was conducted in areas with low species diversity, and it was difficult to identify organism species with sufficient accuracy.
In response to this issue, Professor Kondoh and his fellow researchers developed a technology to detect fish species, in which parts of a trace amount of DNA collected from seawater that identify fish species are selectively amplified, and then these parts are analyzed with the latest equipment to read out their DNA base sequence to determine their species. By filtering a bucket of water or several litters of water taken from a specific sea area and simply extracting and analyzing the DNA remaining on a piece of filter paper, all the fish species living in a specific water area can be comprehensively detected. This method paved the way for conducting monitoring on a scale that had never been achieved by the conventional methods. It is no exaggeration to say that it will bring about an era of big data to biodiversity monitoring.
However, there was still a problem. The detectability of this method had not actually been verified in a sea area that was abundant in fish fauna, due to the absence of reliable comparison data such as visual observation data.

Collecting water samples. A field survey is completed simply by collecting water.
(from https://www.jst.go.jp/pr/announce/20170112/index.html)
Demonstrating high detectability in a rich sea biota
In Maizuru Bay, which faces the Sea of Japan and is located in the northern part of Kyoto Prefecture, since 2002 to the present, Reiji Masuda, Associate Professor at Kyoto University, has collected fish biota data by conducting an underwater visual survey every two weeks. This series of underwater visual surveys is one of the extremely rare cases of marine surveys that have been conducted in such a way as to comprehensively detect fish and obtain thorough observation data on fish biota.
The research group led by Professor Kondoh conducted environmental DNA survey in this sea area on June 18, 2014. As a result of environmental DNA metabarcoding using samples collected at a total of 47 locations, a total of 128 types of fish DNA were detected.
Meanwhile, visual observation data collected from April to August, especially in June (from a total of 140 times of underwater surveys), was compiled, as a result of which, it was found that 80 fish species were observed in Maizuru Bay during this period. Comparing the two data showed that over 60% of fish species observed during 140 underwater visual surveys were included in the results of the metabarcoding.
The results of the underwater visual surveys included 23 types of rare fish species that had been observed only a few times during the 14 years. This indicates that, with exception of these fish species sometimes present in Maizuru Bay depending on the year, by using metabarcoding, as much as 80% of the species that had been observed in the past visual surveys were detected in just one day.
Additionally, in this survey, more than 20 types of fish species that had never been visually observed were detected. Among them, some species of fish larvae that were likely to be overlooked in visual observation were involved. In addition, near the mouth of the river, DNA of freshwater fish was detected, and near the fishing port, DNA of fish that had landed there was also detected. These results show the high level of detectability using this method. It was also revealed that the environmental DNA metabarcoding method for fish survey that was developed in this research and development project makes it possible to comprehensively detect fish communities in a field survey.

Maizuru Bay (from https://www.jst.go.jp/pr/announce/
20170112/index.html)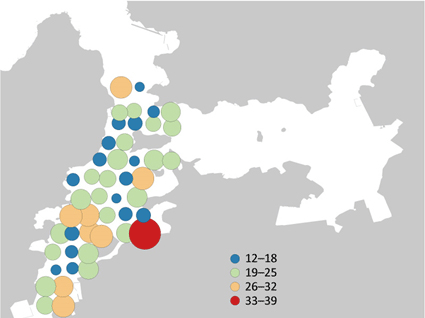
Number of fish species detected by environmental DNA metabarcoding at water sampling points in Maizuru Bay
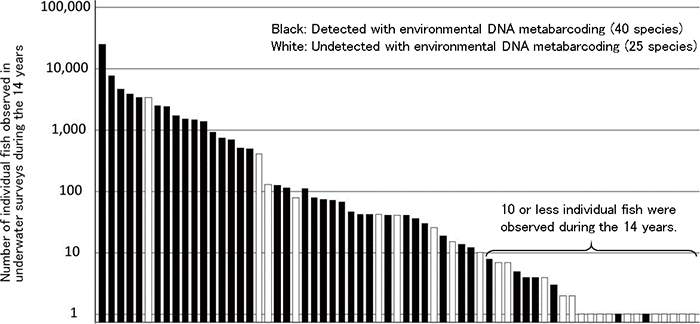
Among the species observed in the underwater visual surveys, the species that were also detected by environmental DNA metabarcoding are shown in the figure. Each vertical axis shows the number of individual fish for each species observed in the visual surveys conducted during the 14 years. The black bars represent species that were also detected by environmental DNA metabarcoding (40 species), while the white bars represent those that were not detected (25 species). In the visual surveys, 80 species were observed. Among those, the graph covers 65 fish species, with the exception of species that cannot be detected by the metabarcoding technology due to system-related reasons. (from https://www.jst.go.jp/pr/announce/20170112/index.html)
Addressing the challenge of surveying the invasion of foreign species
Through this research, it was found that even in a water area with many different fish species in waters around Japan, by simply collecting water samples and analyzing them afterward, research- ers can obtain results that are comparable to those obtained from the series of visual surveys spanning a long period of 14 years. Environmental DNA metabarcoding makes it possible to conduct monitoring of fish communities at "multiple locations" with "high frequency," which was difficult to realize with the conventional survey methods. It is expected that this method will make it possible to conduct the monitoring of the invasion of foreign species and the expansion of their distribution, which have posed a serious problem. Also, it is expected to be used in inaccessible water areas such as deep-sea areas, underground lakes, danger- ous polluted water areas and protected waters where collecting living organisms is prohibited.
