Research Results
Can we edit the genomi information of living organisms?
Resolving the molecular structure of "DNA scissors" FY2018

- Hiroshi Nishimasu (Assistant professor, Graduate School of Science, The University of Tokyo)
- "Sakigake" (PRESTO)
- Structural life science and advanced core technologies for innovative life science research "Development of new genome-editing tools" Researcher (2013-2016)
Rapid evolution of genome-editing technologies
One of the hottest topics in the life science field today is "CRISPR- Cas9". Assistant Professor Hiroshi Nishimasu and his collaborators were the first in the world to report the 3D structure of the Cas9-RNA-DNA complex. Their work was performed as part of the JST "Sakigake" (PRESTO) program, which is aimed at "promoting individual research to nurture the seeds of future innovation and organizing unique, innovative networks". In addition, they have reported a series of world-leading research outcomes, such as the structure-based development of new Cas9 variants and the visualization of the dynamics of Cas9-mediated DNA cleavage.
Cas9 is a bacterial protein that acts on DNA in a scissor-like manner. This cleaving function of Cas9 was biochemically characterized in 2012. Since Cas9-based genome editing techniques are quick and simple, they are used in a wide range of medical applications and basic life sciences for many purposes, such as the production of "knockout animals" (animals lacking certain gene functions) and genetically engineered crops.
Cas9 associates with a guide RNA (an RNA molecule that brings Cas9 to a cleavage site in the DNA target) and then cleaves the target DNA. However, the molecular mechanism of the RNA-guided DNA cleavage by Cas9 was unclear.
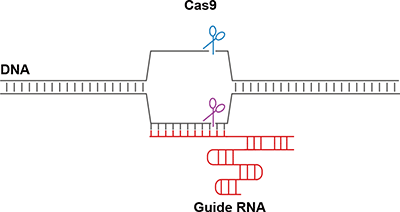
Mechanism of RNA-guided DNA cleavage by Cas9
The first glimpse of the "DNA scissors"
Understanding the action mechanism of Cas9 would lead to the development of more effective genome-editing technologies, Assistant Professor Nishimasu and his collaborators set out to elucidate the mechanism of the Cas9-mediated DNA cleavage.
First, they sought to solve the crystal structure of Cas9 from Streptococcus pyogenes (SpCas9), which is widely used in genome editing, in complex with the guide RNA and its target DNA. After many trials and errors, they crystallized the SpCas9-guide RNA-target DNA complex, obtained X-ray diffraction data at "SPring- ", a synchrotron radiation facility in Hyogo, and determined the crystal structure of the ternary complex. In February 2014, they became the first in the world to report the RNA-guided DNA cleavage mechanism of Cas9. Their crystal structure revealed that Cas9 recognizes a double helix formed by the guide RNA and the target DNA, and the scissor-like parts of Cas9 are located in appropriate positions to cleave the target DNA. This was reported a year after the project started.
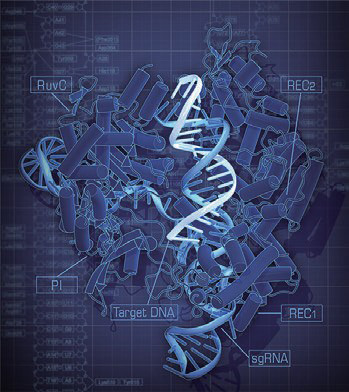
Crystal structure of the SpCas9-guide RNA-target DNA complex
Numerous research teams around the world are working in the CRISPR-Cas field, and research projects are often competitive. Indeed, a research team in the U.S.A. published the Cas9 structure just one week before Dr. Nishimasu and his team published their structure. The rival team solved the structure of the Cas9 protein without the bound nucleic acids (RNA and DNA). Dr. Nishimasu and his team also succeeded in crystallizing Cas9 alone, but surmised that others probably had crystals as well. Therefore, they focused on the ternary complex structure. This decision was critical and helped them to become the first to visualize the molecular structure of the "DNA scissors".
Successful development of new "scissors"!
Although SpCas9 is widely used for genome-editing applications, it is a large molecule that cannot be easily packaged into viral vectors for delivery to target cells. In 2015, it was reported that the Cas9 derived from Staphylococcus aureus (SaCas9) is smaller than SpCas9, and can be packaged into viral vectors more efficiently. Dr. Nishimasu and his team elucidated the crystal structure of the SaCas9-RNA-DNA complex, revealing that SaCas9 has a more streamlined structure as compared to SpCas9.
Based on the structural information, they also succeeded in altering the molecular structure of Cas9 to develop new genome-engineering tools, such as a transcriptional activator and a drug-tunable nuclease.
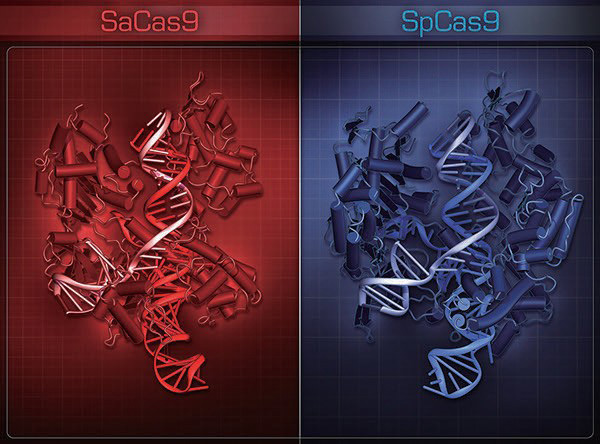
Structural comparison of SaCas9 and SpCas9
Cpf1 (Cas12a), a DNA-cleaving enzyme that differs from Cas9, was discovered in 2015. Dr. Nishimasu and his team determined the crystal structure of the Cpf1-RNA-DNA complex, providing the first insights into the Cpf1-mediated DNA cleavage mechanism. A structural comparison of Cas9 and Cpf1 revealed striking differences in the cleavage mechanisms of these two molecular "scissors". They also successfully developed new genome-editing tools, based on the structural information about Cpf1.
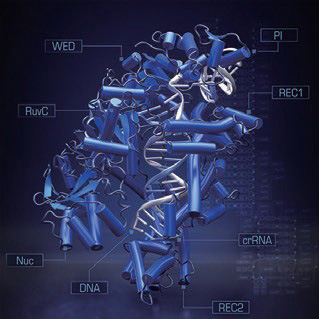
Crystal structure of the Cpf1-guide RNA-target DNA complex
The moment when the "scissors" cut the DNA was captured on video!
Comparisons of the crystal structures of Cas9 alone, the Cas9-RNA binary complex, and the Cas9-RNA-DNA ternary complex suggest that Cas9 undergoes structural changes during the DNA-cleaving process. These crystal structures are, however, static snapshots capturing the moments during the dynamic DNA-cleaving process, and the structural dynamics of Cas9 remained unclear. Dr. Nishimasu and his team therefore teamed up with Associate Professor Noriyuki Kodera and his colleagues at Kanazawa University, in an attempt to visualize the dynamics of Cas9 using high-speed AFM*. In 2017, they successfully captured the Cas9-mediated DNA cleavage process in a single-molecule video, which tremendously impacted the research community around the world. The detailed action mechanism of Cas9 will provide a foundation for the development of more efficient and precise genome-editing tools.
* High-speed AFM (high-speed atomic force microscopy)
High-speed AFM captures real-time video images of nanometer-size biomolecules in aqueous solutions.
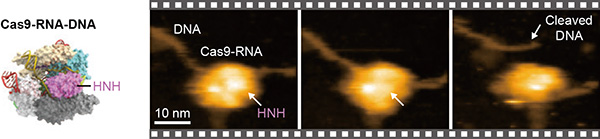
HS-AFM image of the Cas9-guide RNA complex cleaving a target DNA. The part functioning as the "DNA scissors" (the HNH domain) is indicated.
