The research group led by Professor Nobuo Noda of Hokkaido University reconstituted the membrane invagination process required to form autophagosomes(1) in vitro for the first time, showing that this process is mediated by Atg8 protein(2) that plays a central role in autophagy, and a group of enzymes that carry out its lipidation reaction. Autophagy is a mechanism that breaks down and reuses harmful or unwanted substances in cells. It was previously known that Atg8 and the E1, E2, and E3 enzymes responsible for its lipidation play a central role in autophagy, but their role in autophagosome formation, especially in the process of forming its shape, was poorly understood.
In in vitro experiments, the group found that membranes invaginate when all lipidated Atg8 and E1-E2-E3 enzymes are present. At this time, high-speed atomic force microscopy(3) and nuclear magnetic resonance(4) analysis revealed that these proteins form flexible, higher-order complexes on membranes through intrinsically disordered regions(5). Furthermore, yeast experiments confirmed that the E1 enzyme, whose intracellular localization was previously unknown, also localized to the membrane during autophagosome formation along with Atg8 and the E2 and E3 enzymes. These results show that these proteins work together to form autophagosomes.
This study sheds light on the new mechanism of autophagosome formation and is expected to lead to the development of drugs that specifically regulate autophagy.
The research team was consisted with Dr. Noda, Senior Researcher, Tatsuro Maruyama and Postdoctoral Researcher, Mohammed Jahangir Alam of the Institute of Microbial Chemistry in collaboration with the group of Dr. Hitoshi Nakatogawa (Professor, Tokyo Institute of Technology).
This result was obtained from JST Strategic Basic Research Programs team-based research area CREST: "Spatiotemporal dynamics of intracellular components" Research topic: "Autophagy dynamics driven by multi-level higher-order structural components." In this area, we aim at an integrated understanding of cells by observing and measuring the dynamics of higher-order structures in cells, from supramolecular complexes to organelles and membrane-free organelles, and analyzing their functional correlations.
(1) Autophagosome
A bilayer organelle that is newly created in the cytoplasm when autophagy is induced. Everything surrounded by autophagosomes (such as various proteins and mitochondria) is transported to lysosomes (vacuoles in the case of yeast), where they are degraded by the action of a group of degrading enzymes.
(2) Atg protein
It is the name of a group of proteins involved in autophagy identified in yeast, and more than 40 have been reported to date. They are named roughly sequentially in the following formula: "Atg" + the number they were identified. Of the Atg proteins, 19 are important for autophagosome formation during starvation.
(3) High-speed atomic force microscope
This is a microscope that visualizes the shape of molecules based on the atomic force acting between the probe and the sample, and can observe biological molecules such as proteins moving in solution with spatial resolution of nanometers and temporal resolution of subseconds.
(4) Nuclear magnetic resonance
Atomic nuclei that are placed in a strong magnetic field interact with electromagnetic waves of a frequency (resonant frequency) corresponding to the nature of the atomic nucleus and the surrounding environment. Nuclear magnetic resonance is a spectroscopic method that noninvasively acquires information on the structure and properties of proteins and other substances by viewing their electromagnetic waves as signals.
(5) Intrinsically disordered regions
A flexible region that exists in the structure of proteins and does not take a fixed three-dimensional structure. It is involved in various cellular processes through interactions with other proteins and nucleic acids.
-
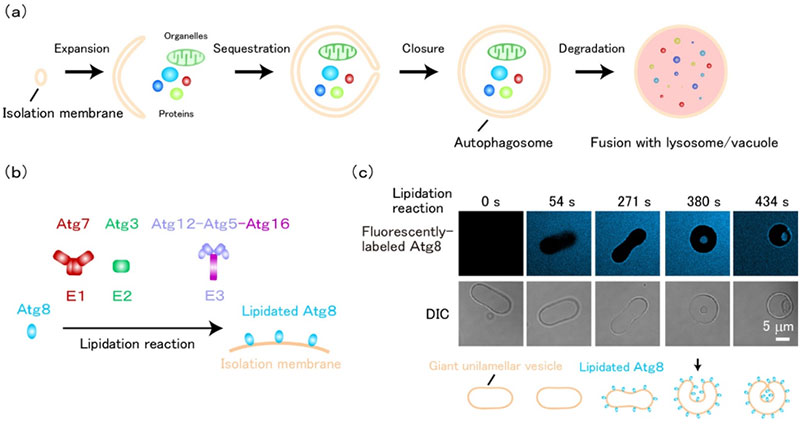
Fig1. Membrane invagination induced by Atg8-E1-E2-E3 - (a) Autophagosomes are formed by membrane expansion and invagination, during which intracellular proteins, organelles, etc. are sequestered into autophagosomes. The sequestered materials are degraded upon fusion of autophagosomes with lysosomes/vacuoles.
- (b) Lipidated Atg8, which is abundant in isolation membranes, is formed by Atg7 (E1), Atg3 (E2), and Atg12-Atg5-Atg16 complex (E3). This enzymatic reaction is called the lipidation reaction.
- (c) When Atg8 lipidation reaction is performed on a prolate-shaped unilamellar membrane, the membrane morphology changes from a prolate shape to a spherical shape with in-bud. (Scale bar: 5 micrometers) ©Nobuo Noda
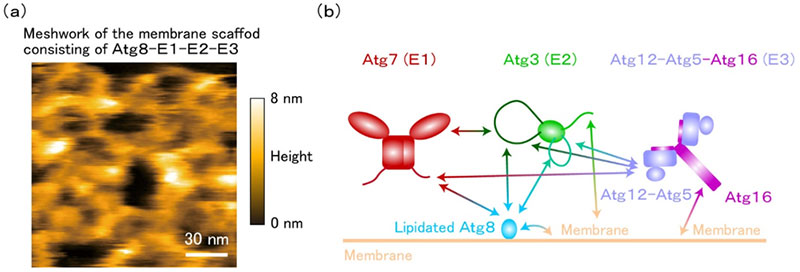
- (a) Lipidated Atg8-E1-E2-E3 forms a membrane scaffold that is flexible and mesh-like (Scale bar: 30 nanometers).
- (b) Lipidated Atg8-E1-E2-E3 membrane scaffold is constructed by multivalent weak protein-protein interactions mediated by intrinsically disordered regions and protein-membrane interactions. ©Nobuo Noda
Program Information
- JST CREST
- Research Area “Spatiotemporal dynamics of intracellular components”
- Research Theme “Autophagy dynamics driven by multi-level higher-order structural components”
Journal Information
Jahangir Md. Alam, Tatsuro Maruyama, Daisuke Noshiro, Chika Kakuta, Tetsuya Kotani, Hitoshi Nakatogawa and Nobuo N. Noda, “Complete set of the Atg8–E1–E2–E3 conjugation machinery forms an interaction web that mediates membrane shaping” Nature structural & molecular biology. Published online December 06 2023, doi: 10.1038/s41594-023-01132-2
Contact
-
[About Research]
Nobuo Noda
Professor, Institute for Genetic Medicine, Hokkaido University
Tel:+81-11-706-5069 Fax:+81-11-706-7826
E-mail: nnigm.hokudai.ac.jp
-
[About Program]
Mutsuko Yasuda
Department of Strategic Basic Research, JST
E-mail: crestjst.go.jp
