Progress Report
Comprehensive Mathematical Understanding of the Complex Control System between Organs and Challenge for Ultra-Early Precision Medicine[2] Experimental Approach to Complex Control System between Organs
Progress until FY2024
1. Outline of the project
Our R&D Item has the following three objectives:
- Obtaining pre-disease datasets and registering them in the pre-disease database (GakuNin RDM).
- Conducting pharmaceutical and medical research on pre-disease states using both biological methods and mathematical methods such as the DNB theory and the control theory.
- Social implementation of ultra-early medical intervention during the pre-disease states.
2. Outcome so far
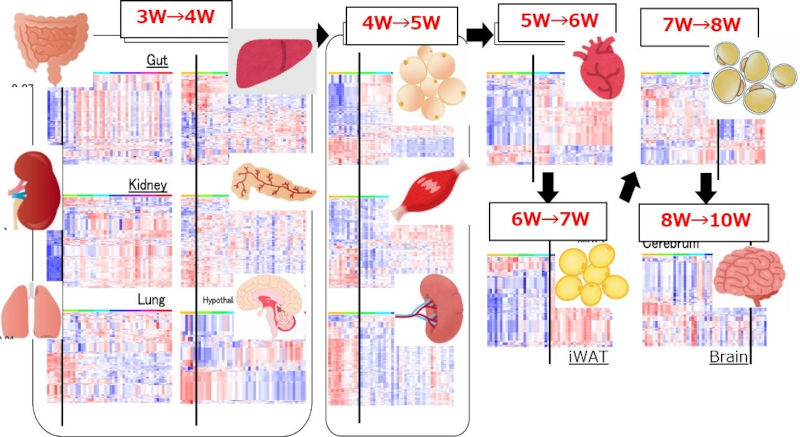
① Acquisition of pre-disease datasets and inter-organ network analysis in metabolic syndrome mice
To elucidate the inter-organ networks, multi-organ and multi-time point gene expression data were obtained for two different types of metabolic syndrome mice and registered in the pre-disease DB.
First, preliminary analysis of gene expression data for 13 organs and 8 time points in spontaneous metabolic syndrome mice (TSOD mice) revealed that the timings of gene expression changes differed between organs, and that the changes appeared to spread from one organ to another. Next, DNB analysis was performed on the gene expression data for the 13 organs of the high-fat metabolic syndrome mouse (C57BL/6 mouse) at 16 time points (Figure 1). The peaks of gene expression fluctuation differed between organs, and it was observed as if the fluctuation was propagating through the organ network before and after the onset of metabolic syndrome.
② Identification of novel therapeutic targets for metabolic syndrome using DNB and control theories
DNB analysis of adipose tissues from TSOD mice identified 147 DNB genes. Collaboration with Prof. Imura's group at narrowed these to eight genes via control theory. Metabolic function analysis in Drosophila revealed Vasa/DDX4 as a factor that suppresses male-specific increased starvation resistance upon high-fat diets (Cells, 2023, 2023).
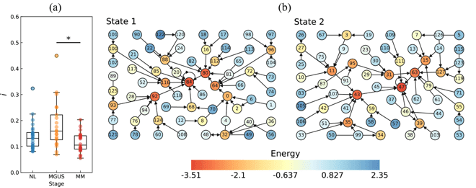
③ Detection of precancerous state (pre-disease state) of hematopoietic tumors using Raman microscopy
We applied DNB theory and energy landscape analysis (ELA) in collaboration with Prof. Naoki Masuda’s group (State University of New York) to Raman spectra obtained from plasma cells of the bone marrow of patients with hematopoietic tumors. The DNB analysis identified MGUS (precancerous condition) as a pre-disease state in the progression of the multiple myeloma stage (Fig. 2a). The ELA revealed two stable states: the normal stage and multiple myeloma in the progression (Fig. 2b). We published two papers on validating the detection of pre-disease states using Raman spectroscopy, DNB theory, and ELA for human samples. (Int. J. Mol. Sci. 2023, 2024).
④ Biology of DNB genes detected in IBD model
To investigate the biological significance of the 27 DNB genes in inflammatory bowel disease (IBD), we calculated control theory-based DNB intervention scores in collaboration with Prof. Jun-ichi Imura’s group and investigated the expression of DNB genes in patients with IBD using GEO database. Subsequently, we performed interventions in Ifit2 and Wars and found that Ifit2 is involved in the pathogenesis of IBD, whereas Wars may play a protective role against IBD. A patent application has been filed with JST on the basis of these results.
3. Future plans
① and ②: We are working on more detailed single cell analysis and intercellular network analysis using visceral adipose tissues from high-fat diet-induced metabolic syndrome mice and humans. ③: By improving the Raman microscope, we are also developing a clinical examination system that can be applied to detect the pre-disease state of human hematopoietic tumors. ④: The detailed biology of DNB genes, including Ifit2 and Wars as candidate intervening genes will be further investigated in humans and mice.