Research Results
Elucidation of the DNA Repair Mechanism in Chromatin
How Double-Strand DNA Breaks are Repaired in ChromatinFY2025

- KURUMIZAKA Hitoshi (Professor, Laboratory of Chromatin Structure and Function, Institute for Quantitative Biosciences, The University of Tokyo)
- ERATO
- Research Director (2019-2024), KURUMIZAKA Chromatin Atlas
RAD51 initiates repair of double-strand DNA break in chromatin
The research group led by Professor Hitoshi Kurumizaka, Laboratory of Chromatin Structure and Function, Institute for Quantitative Biosciences, The University of Tokyo, has revealed how the DNA repair protein RAD51 identifies double-strand DNA break sites within chromatin and initiates DNA repair.
Eukaryotic genomic DNA wraps around histone octamers to form chromatin. Chromatin is folded for storage in the cell nucleus, but its structure dynamically changes in response to gene expression.
The research group led by Professor Kurumizaka has been advancing research to shed light on the structural dynamics of chromatin and regulation of DNA function, as well as to establish the concept of "chromatin atlas," which maps structural information in the nuclear space.
These results may contribute to advancing the establishment of "chromatin atlas" concept and to explaining how cancer develops due to DNA double-strand break.
How does RAD51 repair broken DNA?
Genomic DNA, the blueprint of living organisms, is regularly damaged*1 by ultraviolet rays and cellular metabolism. In particular, double-strand DNA breaks caused by factors including radiation can lead to cancer, the leading cause of death in Japan. However, a considerable portion of cleaved DNA is repaired by DNA repair mechanisms. RAD51 is known to be a central protein involved in this DNA repair.
In eukaryotes, including humans, genomic DNA is stored in the nucleus by forming chromatin, which is an array of nucleosomes*2. The nucleosome consists of DNA tightly wrapped around histone complexes, and this structure itself interferes with DNA repair. The mechanism by which RAD51 binds to nucleosomes in damaged chromatin and promotes DNA repair was unknown (Fig. 1).
*1 DNA damage
DNA damage includes various types of damage that affect genomic DNA sequence and structure, including base oxidation, abnormal base pairing, and DNA breaks. Double-strand breaks, in which both DNA strands are broken, are caused by radiation such as X-rays, environmental substances, DNA replication stress, and other factors. Since DNA damage can cause serious diseases such as cancer and genetic diseases, living organisms have precise mechanisms for its repair.
*2 nucleosome
Nucleosome is the basic unit of chromatin and is structured with approximately 150 base pairs of DNA wrapped around a histone octamer containing two molecules each of four types of histones (H2A, H2B, H3, and H4).
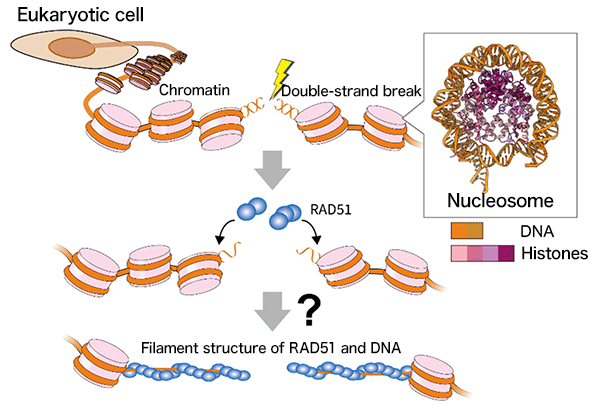
Fig.1 The Enigma of Double-Strand DNA Break Repair in Chromatin by RAD51 The genomic DNA of eukaryotes including humans forms a chromatin structure with nucleosomes as its basic units. Following a double-strand break in genomic DNA, RAD51 accumulates at the site and repairs the DNA. However, the mechanism behind the progression of the reaction in chromatin remained unknown.
Revealing the process by which RAD51 repairs DNA in steps
The research group used cryo-electron microscopy*3 to analyze the structure of complexes comprising RAD51 and nucleosomes that include DNA break ends. As a result, it was discovered that RAD51 forms a ring structure and binds to nucleosomes, recognizes DNA break ends with its ring structure, initiates filament formation by peeling DNA from nucleosomes (Fig. 2). These steps likely correspond to the three processes of accumulation of RAD51 on chromatin, binding to the damaged site, and initiation of DNA repair (Fig. 3).
Meanwhile, the amino-terminal domain of RAD51 (N-terminal lobe domain*4) is unique to eukaryotes, which have chromatin structures, but is absent in prokaryotic RecA (homolog of RAD51), which do not have chromatin structures, and its function was unknown. This research revealed that the N-terminal lobe domain of RAD51 serves an important function in binding to nucleosomal DNA. These results suggest that the RAD51 N-terminal lobe domain was acquired during eukaryotic evolution to facilitate its accumulation on chromatin and repair double-strand breaks. Additionally, the ring structure of RAD51 detects double-strand breaks through the L1 loop*5, which is the active center of double-strand break repair in RAD51 (Fig. 2b, on the right).
These findings revealed that when DNA is broken in chromatin, RAD51 binds to nucleosomes through its N-terminal lobe domain and forms a ring structure to recognize the broken site. This research further illuminated the process in which RAD51 forms a filament structure while pulling DNA away from nucleosomes and initiates DNA repair.
*3 Cryo-electron microscopy
A microscopy in which samples containing proteins are frozen in a cryogenic environment and the ice-embedded samples are observed with electron beams. By taking a large number of micrographs, particle images with various angular information can be obtained, enabling reconstruction of the three-dimensional structure of the protein.
*4 RAD51 amino-terminal lobe domain (N-terminal lobe domain)
The domain at the amino-terminal end of RAD51. RAD51 is composed of two globular domains, including the amino-terminal side and a carboxyl-terminal domain that is homologous to RecA.
*5 L1 loop
A region in the carboxyl-terminal globular domain of RAD51, homologous to RecA, is important for binding to linear DNA.
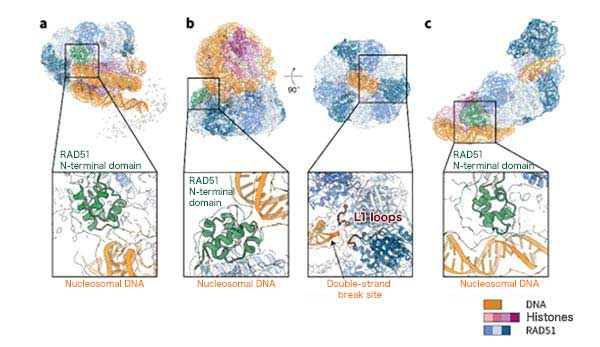
Fig.2 Structures of RAD51 Bound to Nucleosomes (a) RAD51 poised for DNA repair initiation on a nucleosome, (b) RAD51 recognizing a double-strand break site on a nucleosome, and (c) RAD51 initiating DNA repair on a nucleosome. In the structure (b), the L1 loop of RAD51 was bound to the double-strand break site.
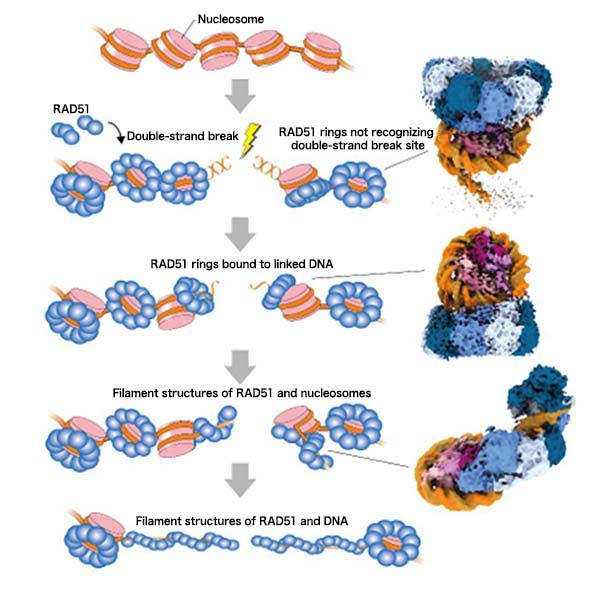
Fig.3 Model of RAD51 Repairing DNA Double-Strand Break in Chromatin RAD51 binds to chromatin in ring structures and recognizes double-strand break sites. Subsequently, RAD51 forms a filament structure by peeling DNA off from the nucleosomes in order to repair the double-strand DNA break. The three structures in the right column correspond to the structures of the RAD51 and nucleosome complexes shown in Fig. 2.
Clarifying the pathogenetic mechanism in cancer and establishing novel treatment methods are expected
It was demonstrated that the N-terminal lobe domain of RAD51 is important for chromatin binding in DNA repair. Amino acid mutations have been found in this domain in many cancer patients. These findings suggest that mutations in the RAD51 N-terminal lobe domain reduce its interaction with nucleosomes during DNA repair and therefore causes carcinogenesis. Further research may contribute to illustrating the mechanism of carcinogenesis caused by mutations in the RAD51 N-terminal lobe domain and to establishing treatment methods.
Professor Kurumizaka and colleagues have been working to shed light on the chromatin atlas, and this research represents one of the outcomes of such efforts. However, fully elucidating the chromatin atlas remains a long-term challenge. A comprehensive understanding of the chromatin atlas could provide fundamental insights into how genetic information is regulated in living organisms and contribute to establishing unprecedented novel treatment methods for related diseases.
- Life Science
- Research Results
- Japanese
