Research Results
Compact and low-cost next-generation diagnostic technology
Development of the fastest detection platform for novel coronavirus (SARS-CoV-2)FY2023

- WATANABE Rikiya (Chief Scientist, RIKEN)
- CREST
- Research Director (2019-2025) of the project, "Single particle technologies of extracellular vesicles for life and medical sciences" in the research area of "Elucidation of biological mechanism of extracellular fine particles and the control system"
Next-generation diagnostic platform based on the world’s fastest detection method “SATORI”
A research group led by Dr. Watanabe, a chief scientist at RIKEN, has successfully developed an automated platform on SATORI: opn-SATORI, a fully automated platform that can identify and rapidly detect viral RNAs*1 derived from the novel coronavirus (SARS-CoV-2) at the single molecule level (Fig.1).
opn-SATORI is based on SATORI, the world's fastest SARS-CoV-2 detection method developed by the Group in 2021, with greatly improved sensitivity and accuracy. It is capable of fully automated quantification of the number of viral RNAs in samples within 9 minutes with sensitivity comparable to PCR and sufficient for diagnosis of COVID-19. It has an accuracy of more than 98% in positive and variant discrimination when validated in clinical samples. In addition, the running cost is approximately $2 per test, which is comparable to PCR and antigen tests.
*1 viral RNA
RNA is ribonucleic acid. Viruses carry genetic information for self-reproduction in very small particles. DNA and RNA carry that genetic information.
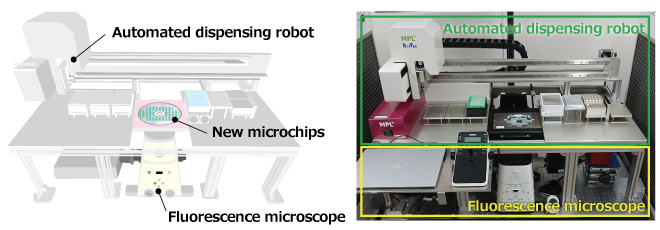
Fig. 1 opn-SATORI platform
Sample preparation, microscopy measurements, virus count quantification, and positive/variant tests were all successfully automated.
Meeting the challenges faced by the conventional SATORI method
The current situation in the diagnosis of novel coronavirus infection (COVID-19) mainly utilizes a method to detect protein antigens (antigen test) and a method to amplify and detect viral RNA (PCR test*2). Each of the two methods can be used for different purposes, such as screening and definitive diagnosis. If infection is suspected, screening is done using the antigen test. The antigen test is suitable for screening because it can quickly and easily detect viruses in about 30 minutes, but its sensitivity and high number of detection errors have been problematic.
Meanwhile, the PCR test, which is used as a definitive diagnosis after screening, employs specialized techniques and devices to purify RNA from specimens, and amplify it further to detect the virus. Although this test is highly sensitive and suitable for definitive diagnosis, it is not easy to quickly analyze a large number of specimens and link them to diagnosis because it takes a minimum of one hour for pretreatment, and detection errors occur due to amplification.
Therefore, the development of a new virus detection method that combines the "high sensitivity" of PCR testing with the "quickness and simplicity" of antigen testing has been eagerly awaited.
In 2021, the research group developed the "SATORI method", the world's fastest way to detect the novel coronavirus, which combines advanced technologies in science and engineering, and has the innovative ability to quickly determine the presence or absence of targeted viral RNA in specimens (Fig. 2).
However, the detection sensitivity of the SATORI method was lower than that of the PCR test, and it was also unable to determine the variants. Moreover, assuming practical applications, a fully automated infection diagnosis process is essential for efficient operation in clinical settings. For these reasons, it has been desired to improve the elemental technology of SATORI, and also to develop a fully automated device.
*2 PCR test
A method that uses polymerase chain reaction to amplify an organism's genes until they are in detectable amounts.
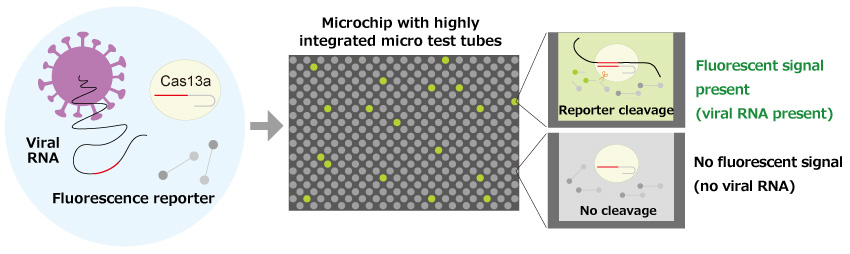
Fig. 2 Unique technology for the world's fastest detection of novel coronavirus (SATORI method)
When the nucleic acid cleaving enzyme CRISPR-Cas13a (Cas13a) is mixed with a fluorescent reporter and the specimen's viral RNA, a complex of viral RNA and Cas13a is formed. Once the complex forms, the enzymatic activity of Cas13a is switched on and the fluorescent reporter is cleaved. Encapsulating the assay mixture in small portions in a microchip array, a fluorescent signal rises within 1 minute for only the test tubes where viral RNA is present. The presence or absence of fluorescence signals in these microchips is binarized, and the number of test tubes with a signal is counted from the digital signal. Since only one molecule of viral RNA is present in a single test tube, the number of test tubes counted corresponds to the number of viral RNA in the sample.
Aiming for low-cost automated detection platform in addition to high-speed and high sensitivity
The research group has greatly improved the elemental technologies of the conventional SATORI method, and has succeeded in developing an opn-SATORI platform that fully automates all steps of the diagnostic test for COVID-19 infection, including sample preparation, microscopic measurement, virus count determination, and positive/variant test. The opn-SATORI platform can identify one viral RNA at the single-molecule level within 9 minutes, and determine the number in a specimen fully automatically (Fig. 3). Its detection sensitivity is 1.4 copies per microliter (1/1,000 mL), which is 1,400 times that of the conventional SATORI method. This sensitivity is comparable to that of the PCR test and is sufficient for the diagnosis of COVID-19.
In addition, a technology to distinguish the single nucleotide*3 variations characteristic of the mutants has also been developed. Along with a positive test, the current platform made it possible to determine known variants, such as the omicron strain. In a validation experiment using COVID-19 clinical specimens, a 98% or higher accuracy response rate was achieved for both positive and variant tests (Fig. 4).
The running cost of the opn-SATORI platform is approximately $2 per test, which is almost the same as that of the PCR and antigen tests. It is expected to become a next-generation diagnostic device for infectious diseases that can accurately diagnose a wide variety of viral infections quickly and inexpensively.
*3 Nucleotide
The main components of RNA (ribonucleic acid) and DNA (deoxyribonucleic acid).
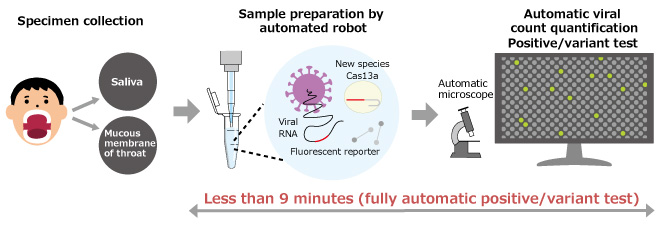
Fig. 3 Fully automated COVID-19 infection diagnosis by opn-SATORI platform
Schematic diagram of infection diagnosis. The whole process of sample preparation, microscopy, viral count quantification and positive/variant tests is automated and can be completed in less than 9 minutes.
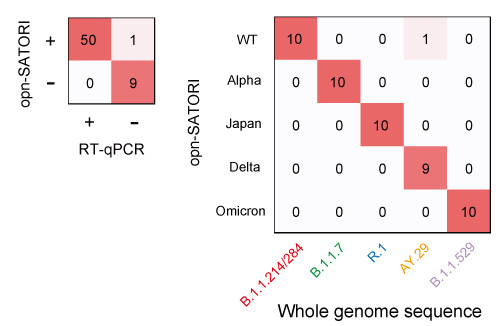
Fig. 4 Validation experiment using COVID-19 clinical specimens
Positive test (right) and variant test (left) results using clinical specimens. Both have an accuracy rate of more than 98%.
Toward next-generation core technologies for quick and wide-range disease diagnosis
The opn-SATORI platform is now being pursued in two directions: one is to expand its application scenarios, and the other is to expand its functions.
To expand the range of applications, the research group subsequently developed "COWFISH (Compact Wide-field Femtoliter-chamber Imaging System for High-speed digital bioanalysis)", a low-cost compact device with a greatly improved detection unit of the opn-SATORI platform. Compared to the regular opn-SATORI platform, COWFISH has been downsized to less than one-fifth the footprint, and has been successfully reduced to about one-thirtieth or less of the cost. In addition, it has achieved quick detection in as little as 3 minutes. The detection sensitivity for SARS-CoV-2 is almost the same as that of the opn-SATORI platform, and in demonstration experiments using clinical specimens, it has achieved a 95% accuracy rate in positive tests.
The downsized and cost-reduced COWFISH will be installed not only in large-scale testing centers but also in city clinics and quarantine stations, and has the potential to become a basic device for conducting point-of-care testing (POCT) (Fig. 5).
In another functional extension, the opn-SATORI platform can be used to detect not only SARS-CoV-2, but also disease biomarkers. In the future, it is also expected to play an important role as a core technology for the next-generation liquid biopsies*4, aiming for early and stratified diagnosis of underlying diseases such as cancer (Fig. 6).
*4 liquid biopsies
A diagnostic method for underlying diseases and infectious diseases based on the analysis of blood and urine, and other humoral samples that are less invasive to the body.
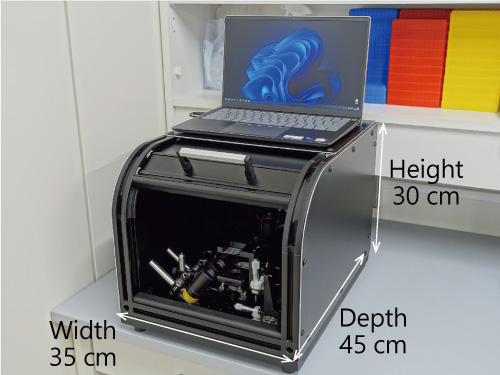
Fig. 5 Compact and low-cost COWFISH
It significantly reduces detection time, and can accurately diagnose a variety of viral infections inexpensively and quickly.
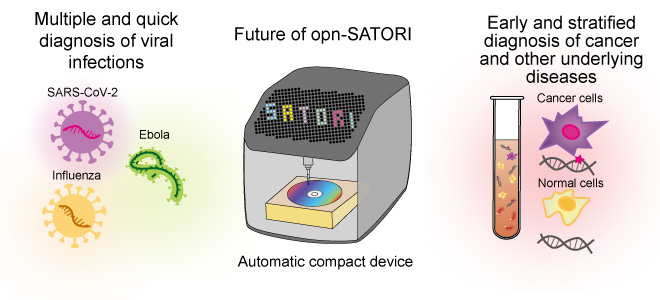
Fig. 6 Future prospects of SATORI method as liquid biopsy
From quick diagnosis of various viral infections to early and stratified diagnosis of underlying diseases such as cancer. Image of the SATORI method as next-generation liquid biopsy.
- Life Science
- Research Results
- Japanese
