Research Results
Also promising as a diagnostic technique for other infectious diseases and cancer
Ultra-sensitive and world's fastest detection of new coronavirusFY2022

- Watanabe Rikiya (Chief Scientist, Molecular Physiology Laboratory, RIKEN Cluster for Pioneering Research)
- CREST
- Research Director (2019-2025), extracellular fine particle area, "Elucidation of biological mechanism of extracellular fine particles and the control system" / "Single particle technologies of extracellular vesicles for life and medical sciences"
Development of the world's fastest detection method, the "SATORI method"
Infection of the new coronavirus has spread, and the number of infected people has continued to increase in Japan as well. The problem was that it took time, effort and money to diagnose the infection of the new coronavirus (SARS-CoV-2).
To cope with this problem, a joint research group led by Research Director Rikiya Watanabe has been promoting interdisciplinary research in science and engineering. The group has succeeded in developing an innovative technique that can identify viral RNA (ribonucleic acid) from SARS-CoV-2 at the "single-molecule" level, and detect it within five minutes. This makes it the world's fastest (*1) new coronavirus detection method, and it was named "CRISPR-based amplification-free digital RNA detection; the SATORI method."
Unlike conventional PCR tests, the SATORI method does not amplify nucleic acids, so there are few errors, and new coronavirus RNAs can be identified and detected one by one within five minutes. As the detection sensitivity is 5 fM (*2), it is possible to detect the amount of viral RNA in a sample from a person infected with the new coronavirus with high sensitivity. Moreover, the running cost is as low as about $9.
These results are expected to be applied as the core technology of next-generation infectious disease diagnostic methods, including the development of ultra-sensitive, rapid diagnostic devices for new coronavirus (COVID-19) and other infections.
*1 As of July 2021
*2 Femtomolar
Written as "fM". 1 fM is 1/1000 trillion [10-15] molar. Molar is molar concentration, which is one way to express concentration. This index is expressed as the number of moles of solute in 1 L of solution with the unit of [mol/L].
A long-awaited new detection method
In the current diagnosis of new coronavirus infection, a method of detecting protein antigens (antigen test), and a method of amplifying and detecting viral RNA (PCR test) (*3) are mainly used. These two are used according to the purpose, such as screening and definitive diagnosis.
If infection is suspected, screening using an antigen test is performed. The antigen test is suitable for screening because it can detect the virus quickly and easily in about 30 minutes. However, the problem is that there are many detection errors due to low detection sensitivity.
On the other hand, in the PCR test, which is used as the next definitive diagnosis after screening, RNA is purified from the sample using special techniques and equipment. Further, the virus is detected through the process of amplification. The PCR test has excellent sensitivity and is suitable for definitive diagnosis. However, pre-processing for detection takes about one hour at the shortest, and detection errors due to amplification also occur. Therefore, it is difficult to quickly analyze and diagnose a large number of samples. Under these circumstances, there was an urgent need to develop a new virus detection method that achieves both the "high sensitivity" of PCR testing and the "rapidity and convenience" of antigen testing.
*3 PCR method
PCR means polymerase chain reaction. The PCR method amplifies the genes of an organism, and increases them to a detectable amount.

Fig. 1 Existing diagnostic techniques for new coronavirus infections
A quick antigen test is used for screening, and a highly accurate PCR test confirms the diagnosis.
The SATORI method enables rapid and highly sensitive analysis in large quantities
The SATORI method is a fusion of advanced technologies related to "single-molecule detection for enzymatic reactions using microchips" by the group of Research Director Watanabe" (*4), and "nucleic acid cleavage enzyme CRISPR-Cas13a (Cas13a) (*5)" by Hiroshi Nishimasu and Osamu Nureki's group at the University of Tokyo. Using a mixture of "Cas13a" that recognizes a specific RNA sequence and a fluorescent reporter (*6) as a biosensor, the presence or absence of target viral RNA in the sample is detected with high sensitivity, high accuracy, and rapidity.
The principle of detecting viral RNA by the SATORI method is as follows (Fig. 2).
*4 Microchip technology
Technology that utilizes semiconductor manufacturing process to fabricate microstructures on chips. In this research, a microchip was created containing about 1 million of the world's smallest level of micro-reactors with a volume of 3 fM.
*5 Nucleic acid cleavage enzyme CRISPR-Cas13a
Many bacteria have an adaptive immune system called the "CRISPR-Cas system" that is acquired when a foreign substance invades. When the same foreign substance invades again, this immune mechanism in the previous memory is activated, and the foreign substance is removed. CRISPR-Cas13a is one of the CRISPR-Cas, and is activated when it forms a complex with guide RNA. When activated, it cleaves not only the target RNA but also the surrounding ssRNA non-specifically.
*6 Fluorescent reporter
A fluorescent functional molecule for detecting a complex of target RNA and Cas13a. This molecule consists of 5nt oligo-RNA whose 5' and 3' ends are labeled with fluorophore and quencher, respectively. In the normal state, fluorescent signal is quenched. However, when oligo-RNA cleaved by Cas13a, which is activated by target RNA, the fluorophore and quencher are physically separated, resulting in the emission of a fluorescent signal.
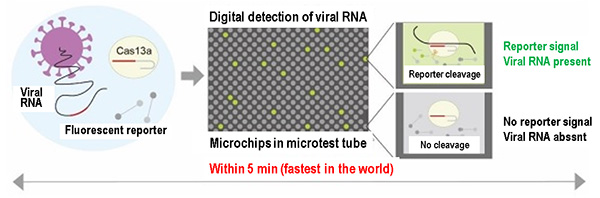
Fig. 2 Principle of digital detection of viral RNA by SATORI method.
A science and engineering collaboration has developed the world's fastest, ultra-high sensitivity system.
1) Mix viral RNA in a mixture of nucleic acid-cleaving enzyme Cas13a and the fluorescent reporter. A complex of viral RNA and Cas13a is then formed.
2) When the complex is formed, the enzymatic activity of Cas13a is switched on, and the fluorescent reporter, in which a fluorescent group and a quenching group are connected, is cleaved.
3) Encapsulate the mixture into femto-liter (fL, 1fL is 1/1000 trillion liters) size micro-reactors . Due to the robust cleavage activity of Cas13a, the fluorescence signal is greatly increased within one minute only in micro-reactors in which viral RNA is present.
4) Count the number of micro-reactors with a fluorescence signal. The number of micro-reactors counted corresponds to the number of viral RNAs in the sample. The volume of each micro-reactor is almost the same as that of E. coli. Viral RNA can be identified and detected at the single-molecule level.
By using the SATORI method, viral RNAs can be identified and detected one by one within five minutes (Fig. 3a). The detection sensitivity is 5 fM (*1). In terms of the amount of viral RNA, this is about 103 per microliter (μL, 1 μL is one-millionth of a liter) (Fig. 3b). This sensitivity is 10 to 100 times higher than that of the conventional antigen test method (104 to 105 copies/μL), and 10 to 100 times lower than that of the PCR test (10 to 102 copies/μL).
However, the amount of viral RNA in the sample of a SARS-CoV-2 infected person is 103 to 106 copies/μL. From this, it can be seen that the SATORI method satisfies the sensitivity required for diagnosing infection with SARS-CoV-2.
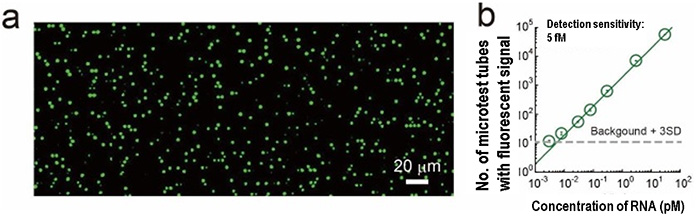
Fig. 3 Example of digital detection example by SATORI method
(a) Fluorescence image of microchips by the SATORI method. Each bright spot corresponds to single molecule of viral RNA.
(b) Correlation between the concentration of viral RNA and the number of micro-reactors in which a fluorescence signal was detected. The limit of detection (LoD) is 5 fM (3×103 copies/μL), which is almost sufficient to detect viral RNA from SARS-CoV-2 patients.
Toward next-generation diagnosis of infectious diseases and early diagnosis of underlying diseases
The SATORI method is an innovative technology that can detect viral RNA at the highest speed in the world. The running cost of the SATORI method is about $9, which is almost the same as the PCR test at about $5. In the future, the SATORI method is expected to become a versatile platform that can rapidly and accurately diagnose a variety of viral infections (Fig. 4). In addition, the SATORI method can be also used to detect various disease biomarkers in liquid biopsy(*7), which aims at early/staging diagnosis of underlying diseases such as cancer (Fig. 4).
A patent application has been filed for these research results, and it is planned to carry out research and development with companies that wish to put it to practical use in the future.
*7 Liquid biopsy
A diagnostic method for underlying diseases and infectious diseases based on the analysis of liquid samples such as blood and urine that is less burdensome to a patient.
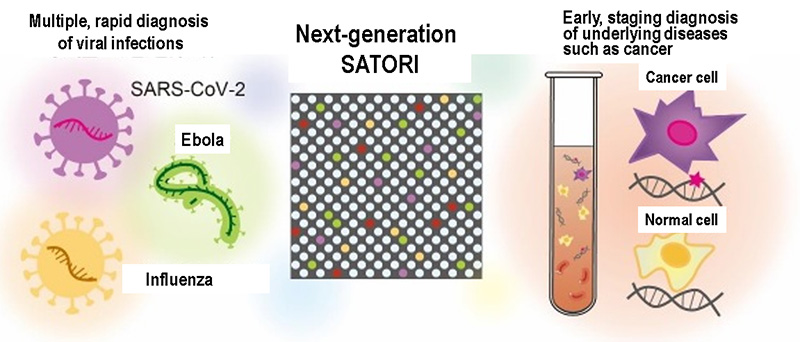
Fig. 4 Future prospects for the SATORI method as liquid biopsy.
Rapid diagnosis ranging from a variety of viral infections to early staging diagnosis of underlying diseases such as cancer. Image of SATORI method as next-generation liquid biopsy.
- Life Science
- Research Results
- Japanese
