- JST Home
- /
- Strategic Basic Research Programs
- /
 CREST
CREST- /
- Research Director/
- Creation of Fundamental Technologies for Understanding and Control of Biosystem Dynamics/
- [Hiroyuki Takeda] Understanding and Predicting 3D Chromatin Dynamics
[Hiroyuki Takeda] Understanding and Predicting 3D Chromatin Dynamics
Research Director

Hiroyuki Takeda
The University of Tokyo
Professor
website
Outline
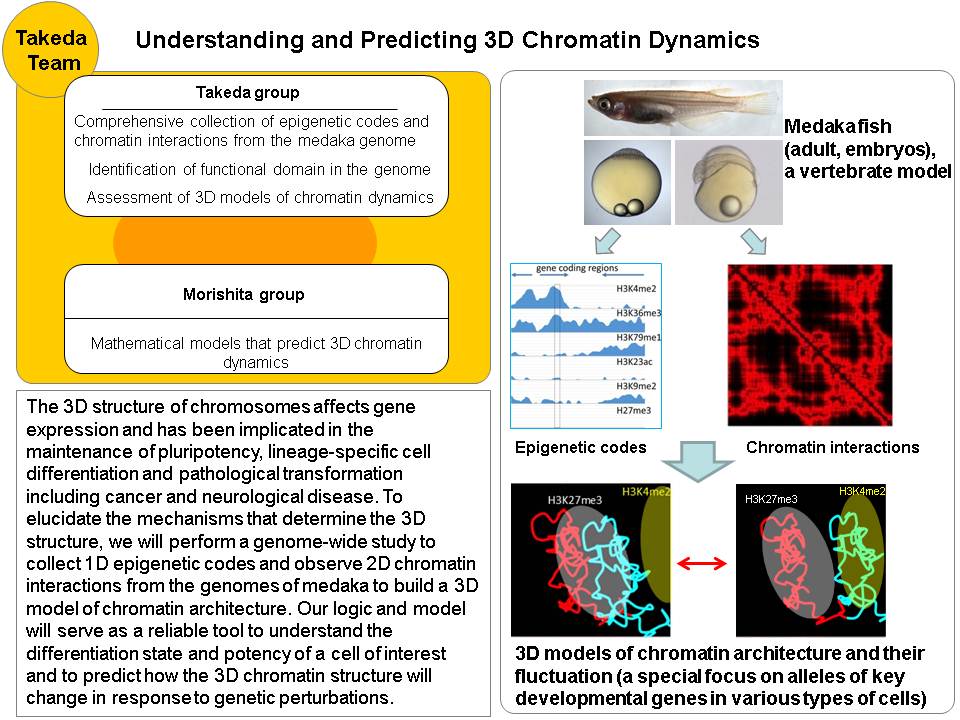
The 3D structure of chromosomes, coiled in the nucleus, affects gene expression and has been implicated in the maintenance of pluripotency, lineage-specific cell differentiation and pathological transformation including cancer and neurological disease. The 3D chromatin structure is thought to be determined by specific DNA binding proteins and epigenetic modifications to DNA (the epigenetic code). However, the mechanisms and factors that determine the 3D structure remain elusive. To address this issue, we will perform a genome-wide study to collect epigenetic codes and observe chromatin interactions from the genomes of medaka fish at various stages of development and cell differentiation. Medaka is an excellent model system for developmental study with a high quality genome sequence available. We will integrate all these data to build a 3D model of chromatin architecture through mathematical modeling using massively parallel computing systems such as the RIKEN K computer. Our logic and model will serve as a reliable tool to understand the differentiation state and potency of a cell of interest and to predict how the 3D chromatin structure will change in response to genetic perturbations.
Collaborators
| Shinichi Morishita | Professor, The Univ. of Tokyo |