- JST Home
- /
- Strategic Basic Research Programs
- /
 ERATO
ERATO- /
- Research Area/Projects/
- Completed/
- IKEDA Genosphere
IKEDA Genosphere

Research Director: Dr. Joh-E. Ikeda
(Professor, The Institute of Medical Science, School of Medicine, Tokai University)
Research Term: 1989-1994
The research conducted in this project was aimed at investigating the genes responsible for the various mental faculties and to explore the spatial arrangement of the human genome, hopefully providing insights into unanswered questions concerning cell functions as well as neural functions.
Research Results
Generation of chromosomal DNA clones: Newly developed methods have enabled chromosomal DNA clones to be generated from undefined regions of chromosomes. The generation of regional human chromosome DNA clones was conducted by laser chromosome microdissection in conjunction with Single Unique Primer-PCR.
Isolation of expressed DNA sequences: A novel approach was developed for isolating expressed DNA sequences (genes) from a defined region of the human chromosomes, which relies on the direct screening of cDNA libraries using region-specific human chromosome DNA clones. This method was applied to the terminal region of the long arm of human chromosome X, on which several causative genes for neurologic and psychomotor diseases have been mapped. It was also applied to the short arm of human chromosome 4 containing the Huntington’s disease.
Ottawa GenoSPHERE laboratory: The Ottawa GenoSPHERE laboratory (located at the University of Ottawa (Canada)) was established. Two main research activities were to generate DNA libraries from microdissected genomic segments of human chromosome 5 and 19, and to isolate the spinal muscular atrophy (SMA) gene region on chromosome 5q13. A strong candidate gene for SMA was isolated.
Spatial territory of chromosomes: The nucleus of eukaryotic cells as been established to be the major compartment in which such functions as chromosome replication and gene expression take place. It is thought that each chromosome occupies a distinct spatial territory in the cell, which affects the processes of cell-specific gene expression.
3-D topography of chromosomes: An imaging system has been developed to reconstruct the 3-D topography of chromosomes, and applied to metaphase cells of the marsupial, in order to analyze the spatial arrangement of its chromosomes. It seems that smaller chromosomes are more likely located in the central region of the chromosome array.
Novel digital imaging system: The digital imaging system was combined with the fluorescent in situ hybridization technique. This combination should reveal the spatial arrangement of chromosomes.
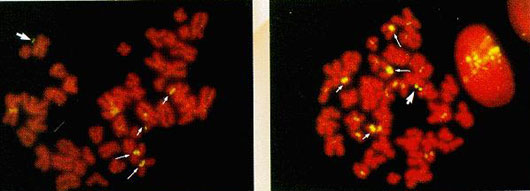
·Hybridization of metaphase spreads from Case 1 with (left) biotin labelled and PCR amplified double minute DNA; FISH signals are observed on double minutes (thick arrow) and on centromeres of D (long, thin arrows) and G (short, thin arrows) group chromosomes (right) alpha satellite probe for chromosomes 13/21; FISH signals are on double minutes (thick arrow), and centromeres of chromosomes 13 (long, thin arrows) and 21 (short, thin arrows).
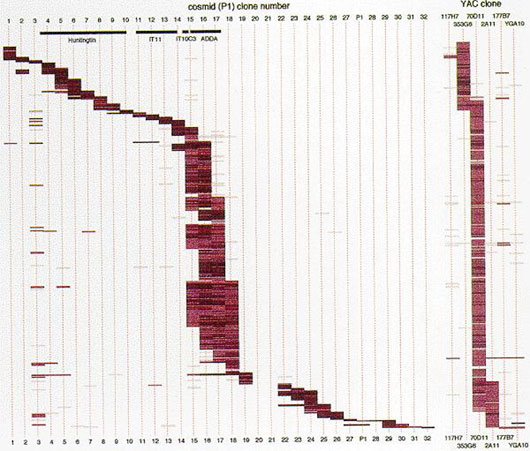
·Ordered map of 578 cDNA clones in 1 Mb of the Huntington’s Disease gene region.
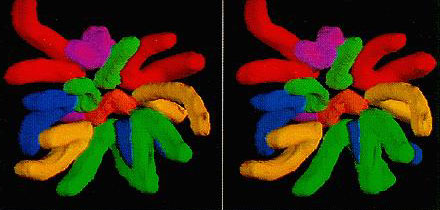
·Stereo gram of the image of the reconstructs of the assigned 3-dimensional chromosome.

















