- JST Home
- /
- Strategic Basic Research Programs
- /
 ERATO
ERATO- /
- Research Area/Projects/
- Completed/
- HORIKOSHI Gene Selector
HORIKOSHI Gene Selector

Research Director: Masami Horikoshi
(Associate Professor, Institute of Molecular and Cellular Biosciences, The University of Tokyo)
Research Term: 1997-2002
The aim of this project was aimed at understanding how multicellular organisms develop during and after embryogenesis with a single set of chromosomes for a variety of cells.
Research Results
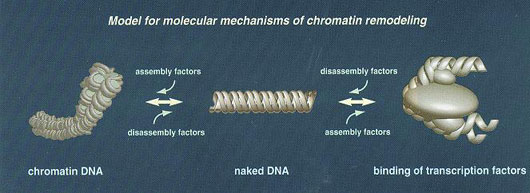
New ‘gene selectors’ identified: A wide variety of techniques were used to identify ‘gene selectors’ in the nucleus associated with the chromatin-coupled transcription process. Many interaction networks were identified. Histone chaperone CIA and MYST-type HAT (histone acetylase) are the most interesting ‘gene selectors’ obtained. In addition to these, about 200 others have been identified.
‘Chromosomal gradient’ of histone accetylation: A pair of HAT and HDAC (histone deacetylase) has been revealed to establish a ‘chromosomal gradient’ of histone acetylation around a chromosome terminus called telomere. In yeast S. cerevisiae, telomeric regions are hypoacetylated at the Lys-16 of histone H4, but hyperacetylated at the same lysine in telomere-distal regions. Sir2, an NAD dependent HDAC, and Sas2, a member of MYST-type HAT family, are responsible for the hypoacetylation or the hyperacetylation, respectively. The loss of hyperacetylation induced by deleting the SAS2 mutation, is accompanied by inactivation of gene expression in telomere-distal regions. The acetylation of Lys16 of histone H4 by Sas2 in telomere-distal regions has been found to function as an anti-silencing mechanism to prevent silenced chromatin to spread from telomeric regions.
Common conserved motif: Another pair of HAT and HDAC, Esa1 and Rpd3, acts competitively in gene regulation. A common and conserved motif in the primary structures of Esa1 and Rpd3 (the ER-motif) was observed. This motif resides near the active center in the both enzymes, and is involved in enzymatic reactions in a similar manner. It has been proposed that there may be a structural similarity between pairs of enzymes that possess opposite functions in the cell.
Cellular roles of ‘gene selectors’: Unicellular organisms, such as yeast and bacteria, were analyzed using electron microscopy in an effort to elucidate the cellular roles of ‘gene selectors’. The most important result obtained by this method is the involvement of ‘gene selectors’, named CIA, MYST-HAT and UNI1, in the cell death regulatory system. Surprisingly, apoptosis-like ‘prototypal active cell death’ has been observed not only in yeast S. cerevisiae, but also in bacteria E. coli using electron microscopy. These findings provided an exploratory cue to understand the unified mechanisms of cell death.
Three-dimensional structures of ‘gene selectors’: Once many ‘gene selectors’ and their interaction networks were identified, their functions, chemical relations and phenotypes were described. In addition to these, the determination of the 3-dimensional structures of particular ‘gene selectors’ is necessary for their characterization at the atomic level. Tertiary structures of the novel ‘gene selectors’, such as CCG1-interacting factor B (CIB) and yeast gankyrin orthologue Nas6, were solved.